Source: Helle Palmø, chief geneticist, DanBred, 15 November 2020, photo credit: EMBL-EBI Training
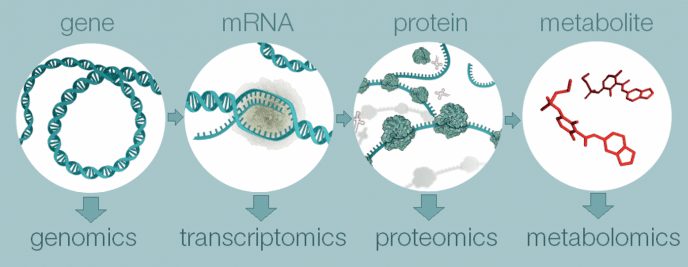
Metabolomic selection can improve genetic gain in difficult-to-measure traits by combining whole-metabolomic data with phenotypic, pedigree, and genomic data. The metabolome is the complete set of metabolites in a sample and forms a link in the chain between DNA and phenotype.
New breeding technologies such as genomic selection, whole-genome sequencing, optimal contribution selection, and gene editing are developing at an increasing pace. Metabolomic selection is an emerging breeding technology based on an exciting new source of information, namely nuclear magnetic resonance (or NMR) metabolomics.
NMR metabolomics measures all metabolites in a sample from an individual. This complete set of metabolites – referred to as whole-metabolomic data – is associated with the level of physiological activity in biological pathways that are initiated at DNA level and culminate in trait expression.
The level of physiological activity is, in turn, regulated by the genes that an individual has inherited from its parents, as well as by cues from its environment. This link between whole-metabolomic data and inherited genes may be exploited to increase the genetic potential for desirable phenotypic traits such as feed efficiency and meat quality in pigs.
Read more
The South African Pork Producers’ Organisation (SAPPO) coordinates industry interventions and collaboratively manages risks in the value chain to enable the sustainability and profitability of pork producers in South Africa.